Example Data in phyloseq
Mon Mar 12 15:05:42 2018
There are multiple example data sets included in phyloseq. Many are from published investigations and include documentation with a summary and references, as well as some example code representing some aspect of analysis available in phyloseq.
Load Packages
library("phyloseq"); packageVersion("phyloseq")## [1] '1.22.3'library("ggplot2"); packageVersion("ggplot2")## [1] '2.2.1'ggplot2 package theme set. See the ggplot2 online documentation for further help.
theme_set(theme_bw())Load Example Data
To load example data into the working environment, use the data() command:
data(GlobalPatterns)
data(esophagus)
data(enterotype)
data(soilrep)Documentation
In the package index, go to the names beginning with “data-” to see the documentation of currently available example datasets.
Try for example
?GlobalPatternsto access the documentation for the so-called “GlobalPatterns” dataset.
Run provided examples
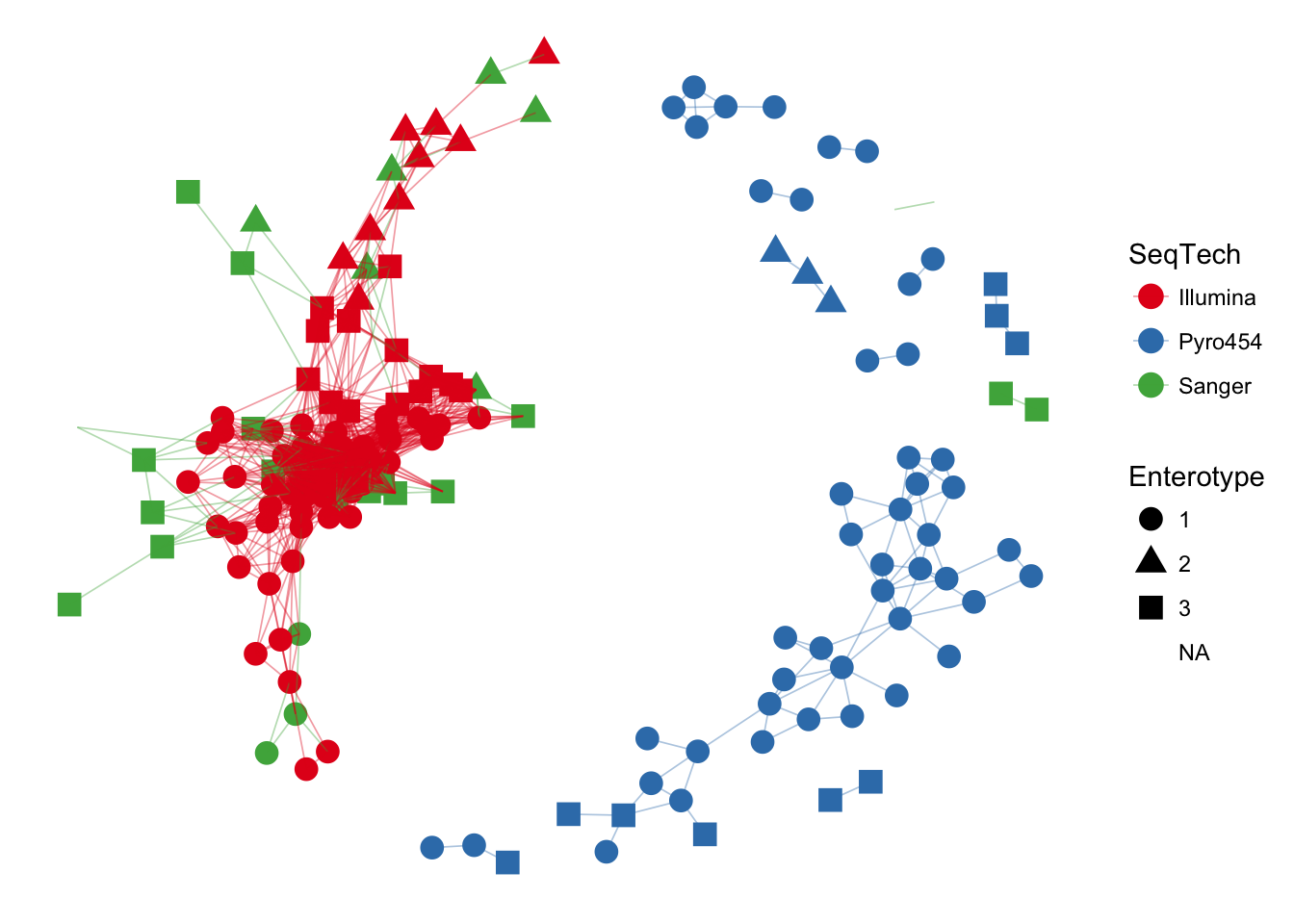
You can also try the examples included with the example data documentation (as well as examples for functions/methods) using the standard example command in R – in this case the examples for the enterotype dataset.
example(enterotype, ask=FALSE)##
## entrty> data(enterotype)
##
## entrty> ig <- make_network(enterotype, "samples", max.dist=0.3)
##
## entrty> plot_network(ig, enterotype, color="SeqTech", shape="Enterotype", line_weight=0.3, label=NULL)## Warning: attributes are not identical across measure variables; they will
## be dropped